[PRESS RELEASE – San Francisco, California, December 31st, 2024]
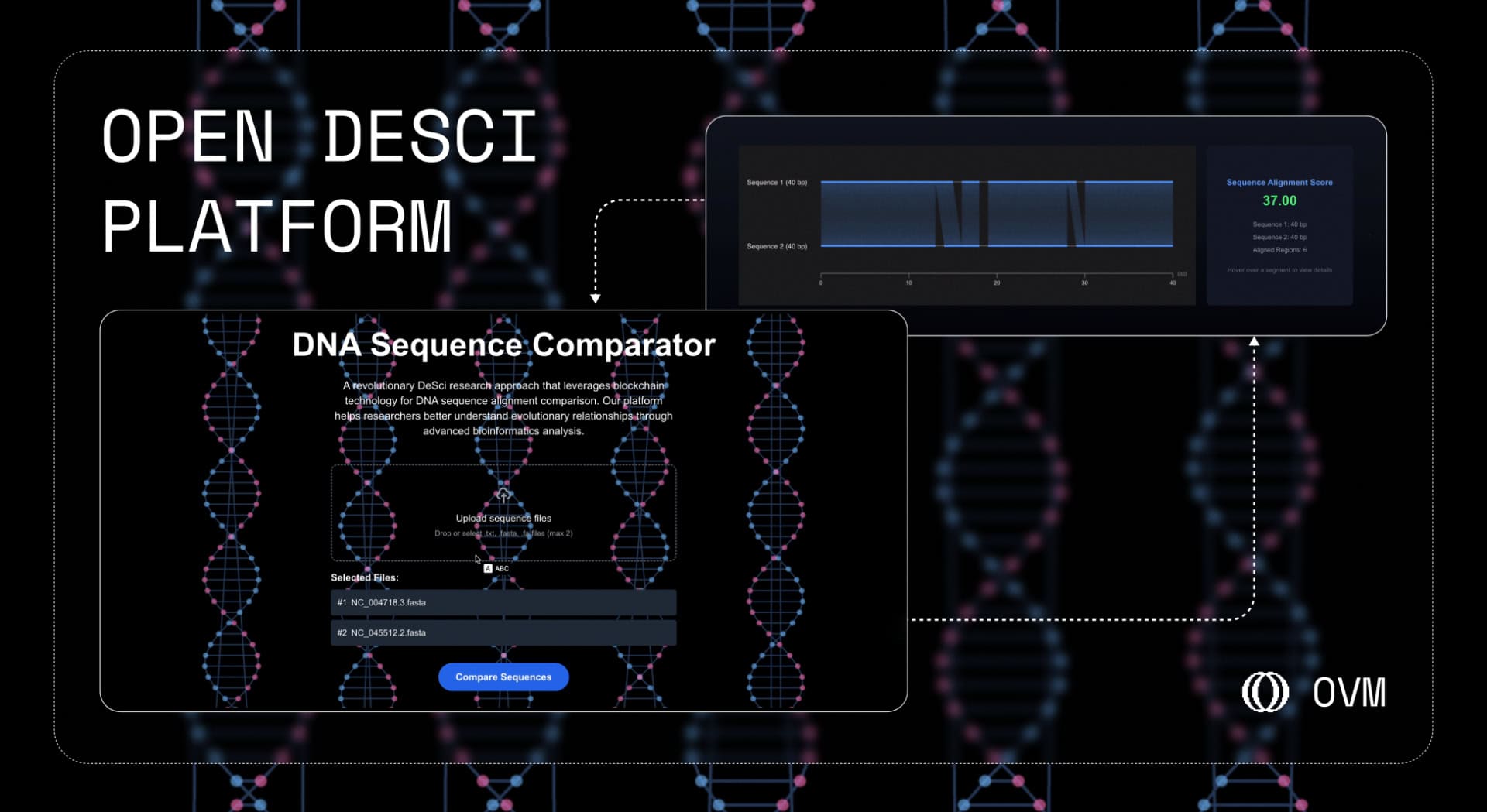
Open (also known as webisOpen on X, collective includes RSS3) is transforming virological research and decentralized science (DeSci) through its innovative Open Virtual Machine (OVM). This technology, integrated with the Compute Wormhole, facilitates the on-chain analysis of genetic similarities between two viruses: SARS-CoV-2 (sequenced in Wuhan, 2019) and SARS-CoV (sequenced in Toronto, 2003).
This analysis is performed fully on-chain using Open’s Open Virtual Machine (OVM) within a Trusted Execution Environment (TEE) (together known as Open’s Compute Wormhole) utilizing and bridging decentralized compute from Hyperbolic Labs, to ensure integrity, immutability, determinism, and resistance to tampering. Additionally, OVM guarantees verifiability through on-chain transactions, promoting transparency and trust in the results.
This demo and experiment of Open Desci Platform Alpha and its record on-chain (on Open Chain) has also been released to the public to showcase the power of OVM to the public. Users can check out this link for more information.
The Research Challenge
Decoding the genetic fingerprints of viruses has traditionally required expensive supercomputers and extensive infrastructure—resources often out of reach for many researchers. For years, virologists have been constrained by financial and logistical challenges, forcing them to focus more on acquiring equipment than advancing scientific discovery. Moreover, the reliance on private or centralized computing resources has limited peer reviews, making it difficult to reproduce results and raising concerns about the integrity, reliability, and trustworthiness of the findings.
Entering the OVM Revolution
The Open Virtual Machine (OVM) offers a high-performance, decentralized, and verifiable compute layer, designed to democratize access to advanced computational resources. Its implementation begins with a demonstration and experime
Go to Source to See Full Article
Author: Chainwire